Please visit the new version of LipiOne 2.0 at lipidone.eu.
GENERAL DESCRIPTION
![]() LipidOne, a freely available software that processes lipidomics data to help researchers to find new relationships between lipid profile and the activity of enzymes/genes involved.
LipidOne, a freely available software that processes lipidomics data to help researchers to find new relationships between lipid profile and the activity of enzymes/genes involved.
LipidOne, automatically highlights all qualitative and quantitative changes in lipid building blocks both among all detected lipid classes and among experimental groups. Thanks to LipiOne, the discovered differences in lipid building blocks can be easily linked to the activity of specific enzyme classes and or gene expression.
CITATION
Please, cite our work as:
Roberto Maria Pellegrino, Matteo Giulietti, Husam B R Alabed, Sandra Buratta, Lorena Urbanelli, Francesco Piva, Carla Emiliani, LipidOne: user-friendly lipidomic data analysis tool for a deeper interpretation in a systems biology scenario, Bioinformatics, 2021; btab867, https://doi.org/10.1093/bioinformatics/btab867
CONTACT
For general questions concerning the use of the software please contact Roberto Maria Pellegrino (This email address is being protected from spambots. You need JavaScript enabled to view it.)
For problems with installation, operation, software compatibility contact Matteo Giulietti (This email address is being protected from spambots. You need JavaScript enabled to view it.)
LipidOne is an executable file that does not require installation. Follow the instructions below and try working with our example datasets.
Instruction:
a) For the first time only, the user must install the Gnuplot graphics libraries. They can be downloaded free of charge from: https://sourceforge.net/projects/gnuplot/files/gnuplot/5.4.2;
b) Download LipidOne and place it in a folder of the user's choice.
c) Double click on the icon to start LipidOne.
*) We would like to thank Dmitry Karasik and colleagues for the development of the Perl Prima library (http://www.prima.eu.org/), which we used for the development of the Graphical User Interface of LipidOne.
**) We also thank Thomas Williams and colleagues for their GNUPLOT graphical library (http://www.gnuplot.info/), useful for the creation of the LipidOne bar graphs.
Try and explore some example datasets with LipidOne.
RETINA: Trzeciecka, A. et al. (2019) Comparative lipid profiling dataset of the inflammation-induced optic nerve regeneration. Data in Brief, 2352-3409.
FUNGUS: Lipid profiling of fungus (our internal, unpublished results).
ALGAE: Lipid profiling of algae from Riken Demonstration Files (http://prime.psc.riken.jp/compms/msdial/main.html).
Note: If the lipid annotation of your dataset does not comply with the LSI guidelines*, you can convert your data with LipidLynxX Converter.
---
*) Liebisch,G. et al. (2020) Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. Journal of Lipid Research, 61, 1539-1555.
 Three-step LipidOne workflow
Three-step LipidOne workflow
1) Data upload: the user must select a data matrix (txt or csv tab delimited file) containing the names and concentrations in each lipid sample detected (see example dataset). The accepted lipid nomenclature is the "molecular species level" or "sn-position level" as reported by the Lipidomics Standards Initiative (LSI) Guidelines.
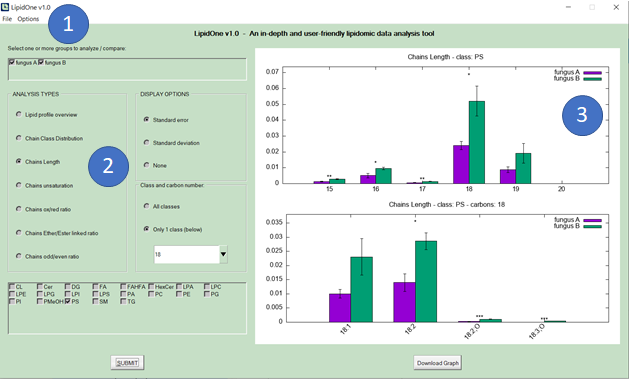
2) Query selection: The user starts to explore the dataset using seven types of analysis and dynamic context-dependent menus.
3) Get results: all graphs and tables can be exported.
GENERAL STATEMENT
LipidOne is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
DISCLAIMER
LipidOne is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
CONTACT
For general questions concerning the use of the software please contact Roberto Maria Pellegrino (This email address is being protected from spambots. You need JavaScript enabled to view it.)
For problems with installation, operation, software compatibility contact Matteo Giulietti (This email address is being protected from spambots. You need JavaScript enabled to view it.)


